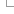Opossum vs Tasmanian devil Synteny results
The syntenic regions between Opossum (Monodelphis domestica, ASM229v1) and Tasmanian devil (Sarcophilus harrisii, mSarHar1.11) were extracted from their pairwise alignment in Ensembl release 102. We look for stretches where the alignment blocks are in synteny. The search is run in two phases. In the first one, syntenic alignments that are closer than 200 kbp are grouped. In the second phase, the groups that are in synteny are linked provided that no more than 2 non-syntenic groups are found between them and they are less than 3Mbp apart.
Configuration parameters
No configuration parameters are available.
Statistics over 242 synteny blocks
| Genome coverage (bp) | Coding exon coverage (bp) | |
|---|---|---|
| Opossum |
Uncovered: 176,811,812 out of 3,600,504,728 |
Uncovered: 2,123,345 out of 35,083,223 |
| Tasmanian devil |
Uncovered: 77,010,353 out of 3,086,657,325 |
Uncovered: 1,797,336 out of 33,291,752 |